Stingrays are the most species-rich venomous cartilaginous fish order. They comprise 218 extant species distributed circumglobally in marine, brackish, and freshwater habitats (Fishbase; Weigmann, 2016).They possess a defensive venom system in form of a retroserrated tail spine covered by venom-secretory cells, mostly located in the lateral grooves and embedded in an epidermal sheet (Pedroso et al., 2007)
Stingrays

Evolutionary background
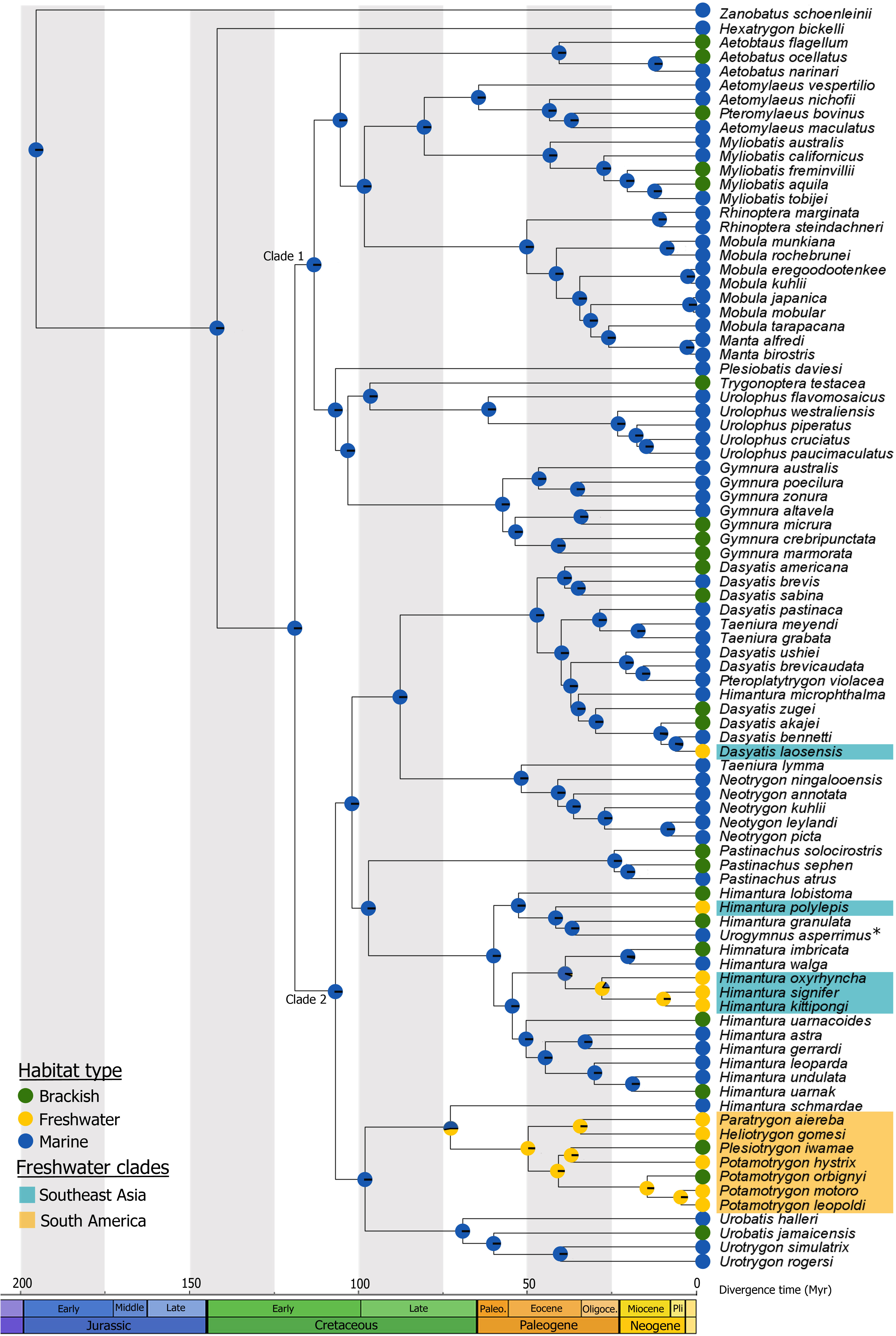
Species richness in freshwater bony fishes depends on two main processes: the transition into and the diversification within freshwater habitats. In contrast to bony fishes, only few cartilaginous fishes, mostly stingrays (Myliobatoidei), were able to colonize fresh water. Respective transition processes have been mainly assessed from a physiological and morphological perspective,indicating that the freshwater lifestyle is strongly limited by the ability to perform osmoregulatory adaptations. However, the transition history and the effect of physiological constraints on the diversification in stingrays remain poorly understood. Herein, we estimated the geographic pathways of freshwater colonization and inferred the mode of habitat transitions. Further, we assessed habitat-related speciation rates in a time-calibrated phylogenetic framework to understand factors driving the transition of stingrays nto and the diversification within fresh water. Using South American and Southeast Asian freshwater taxa as model organisms, we found one independent freshwater colonization event by stingrays in South America and at least three in Southeast Asia. We revealed that vicariant processes most likely caused freshwater transition during the time of major marine incursions. The habitat transition rates indicate that brackish water species switch preferably back into marine than forth into freshwater habitats. Moreover, our results showed significantly lower diversification rates in brackish water lineages, whereas freshwater and marine lineages exhibit similar rates. Thus, brackish water habitats may have functioned as evolutionary bottlenecks for the colonization of fresh water by stingrays, probably because of the higher variability of environmental conditions in brackish water.
Figure shows the ancestral habitat estimation in stingrays. Ancestral habitat estimates (marine, brackish water, and fresh water) obtained by a MuSSE analysis. For details see Kirchhoff et al., 2017
Intraspecific variability
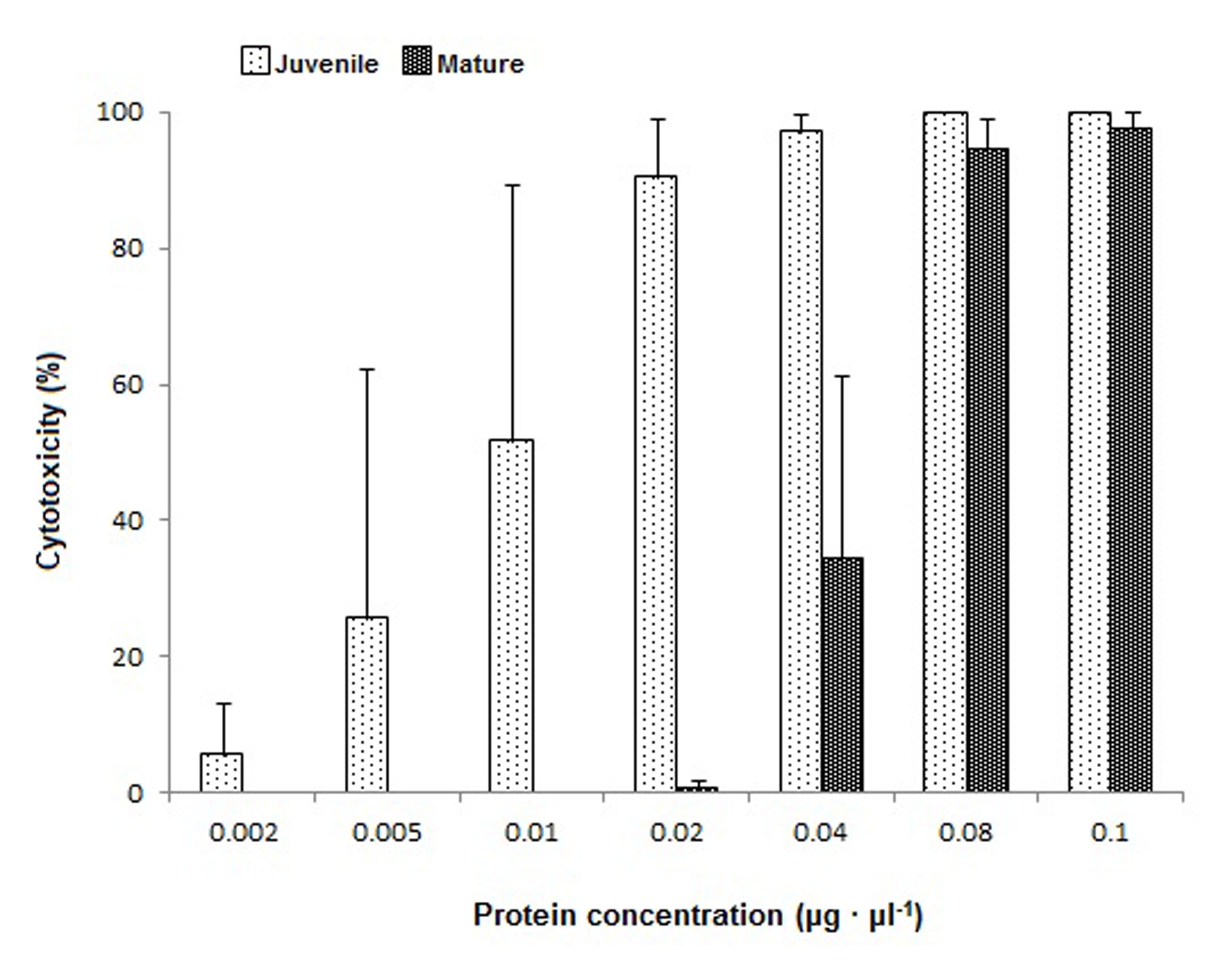
Aquatic venomous animals such as stingrays represent a largely untapped source for venom-based drug development. However, the major challenge for a potential drug development pipeline is the high inter- and intraspecific variability in toxicity and venom composition. As of today, little is known about maturity-driven changes in these traits in stingrays. The present study investigates the differences in toxicity and venom composition in different maturity stages of the freshwater stingray Potamotrygon leopoldi. This species can be found in the Xingú River basin (Brazil), where it mainly feeds on invertebrates, while being predated by other stingrays or large catfishes. P. leopoldi, as commonly known for stingrays, can cause severe injuries with the venomous dentine spine located at its tails. The toxicity of tissue extracts of juvenile and mature specimens was recorded on a myoblast cell culture bioassay. Venom composition and bioactivity of compounds were analyzed with planar chromatography linked to an Aliivibrio fischeri bioassay. Results revealed a decrease in venom toxicity during maturation, but no changes in venom composition. These findings may indicate that toxicity in mature specimens becomes evolutionary less important, probably due to a decrease in predation pressure.
Figure shows the cytotoxic activity of juveniles and mature Potamotrygon leopoldi tissue extracts on mouse myblasts. For details see Kirchhoff et al., 2014
Interspecific variability
The venomous spine tissue of one mature specimen of five marine and freshwater stingray species has been assessed by transcriptomics (1) and three of them further by high-content screening (2). The transcriptomic approach gives us a putative venom composition by annotating the high quality contig (assembled cDNA reads) against ToxProt. The toxin family evaluation of these transcriptomes indicates that the venom of stingray exhibits high dominancy of ‘Translational-controlled tumor proteins’ and Hyaluronidases. The high-content screening, which is a HT cell-based bioassay, gives us a first insight into activity patterns and some indicators for the potential mechanism of action of stingray toxins. Here the effects on HeLa cells indicates that stingray venoms act on the mitochondria, ER, lysosome and disrupts the cytoskeleton.
Figure shows (1) Transcriptomic data: Circos with toxin family-related abundance, expression, functional and candidate data and a Venn diagram for interspecific comparison; and (2) High-content screening: Circos visualizing the cytological profiles induced by stingray venom and the c. 100 reference compounds clustering with them and images of stained cells obtained from the cellomics platform. Coming soon: Kirchhoff et al., in prep
Applicability
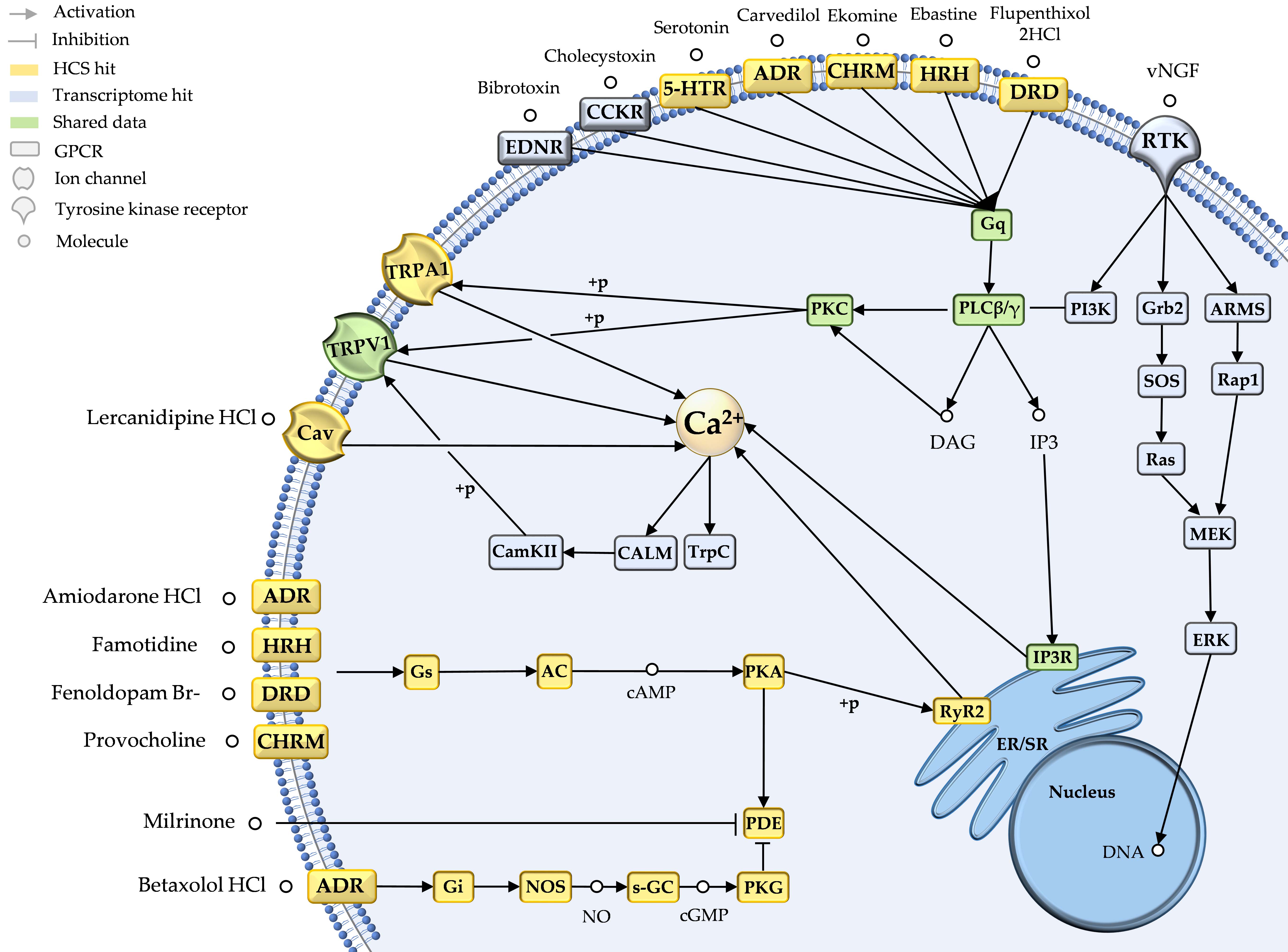
One of the most important barriers for novel natural products in applied fields is their amount and the need for a cost-effective and fast method for the primary assessment of their potential. Therefore, in my newest publication (currently in preparation) we integrated transcriptome and high-content screening (HCS) data on functional level in order to obtain a fast overall prediction of the pharmacological potential of the poorly known stingray venom system. This integration enables the identification of putative candidates in stingray venom and links them to target system, molecular target groups and even induced cellular responses.
Figure shows the exemplary pain-response network constructed from the transcriptome (blue)- and HCS (yellow)-integrated functional data. It shows that putative pain-inducing stingray toxins bind to G-protein coupled receptors and activate intracellular calcium signaling mostly through the IP3 signaling pathway. Coming soon: Kirchhoff et al., in prep
Techniques
- Evolutionary biology: From primer design and PCR to phylogenetic analysis
- Biochemistry: Performance of HPTLC; Introduced into Size Exclusion & Ion exchange FPLC and 2D SDS-PAGE linked to MALDI-TOF performance and data evaluation
- Physiology: Experienced in mouse cell cultivation and based on them viability/cytotoxicity assays; Confident with High-content screening data evaluation and interpretation
- Genetics: Beside phylogenetic-based genetic work, I am experienced in transcriptome data handling and evaluation
- Protein expression: Introduced into FXa recombinant expression in E. coli and Cell free expression





